Secondary structures such as G-quadruplexes (G4s) can form within DNA or RNA. They pose a dramatic risk for genome instability, because due to their stability they can block DNA replication and this could lead to DNA breaks. In certain cancer cells mutations/deletions are observed at G4s, if a helicase that is important for G4 unwinding is mutated. Nevertheless, G4s are also discussed to be functional elements for cellular processes such as telomere protection, transcription, replication, and meiosis.
The Paeschke group uses a combination of genetic, molecular biological and genome-wide approaches to identify and characterize novel G-quadruplex (G4)-interacting proteins. Initial experiments are performed using yeast as a model organism and gained information will be transferred into human cells where results will be linked to human health. They have developed three novel screening techniques that enable us to identify novel G4 interacts. Candidate proteins are further validated in vitro as well as in vivo.
Due to the connection of G4s and cancer the data obtained in the Paeschke lab will not only be important to understand G4 regulation and formation, but will also provide unique knowledge on the impact of G4 structures for genome stability and thereby for human health.
Background: |
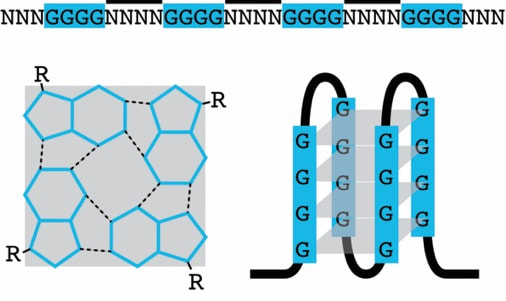
G4s are stable structures that can form in guanine-rich regions of RNA and DNA with specific sequence motifs. Although G4 structures were already discovered 1962 only recently a lot of attention was drawn to the biological function and challenges of G4 structures in vivo. Due to their stability G4 structures are discussed as a novel tool within cells that support and alter different biological processes. We and others have demonstrated, that G4 formation is a dynamic process that differs within the cell cycle but also among different cell types.
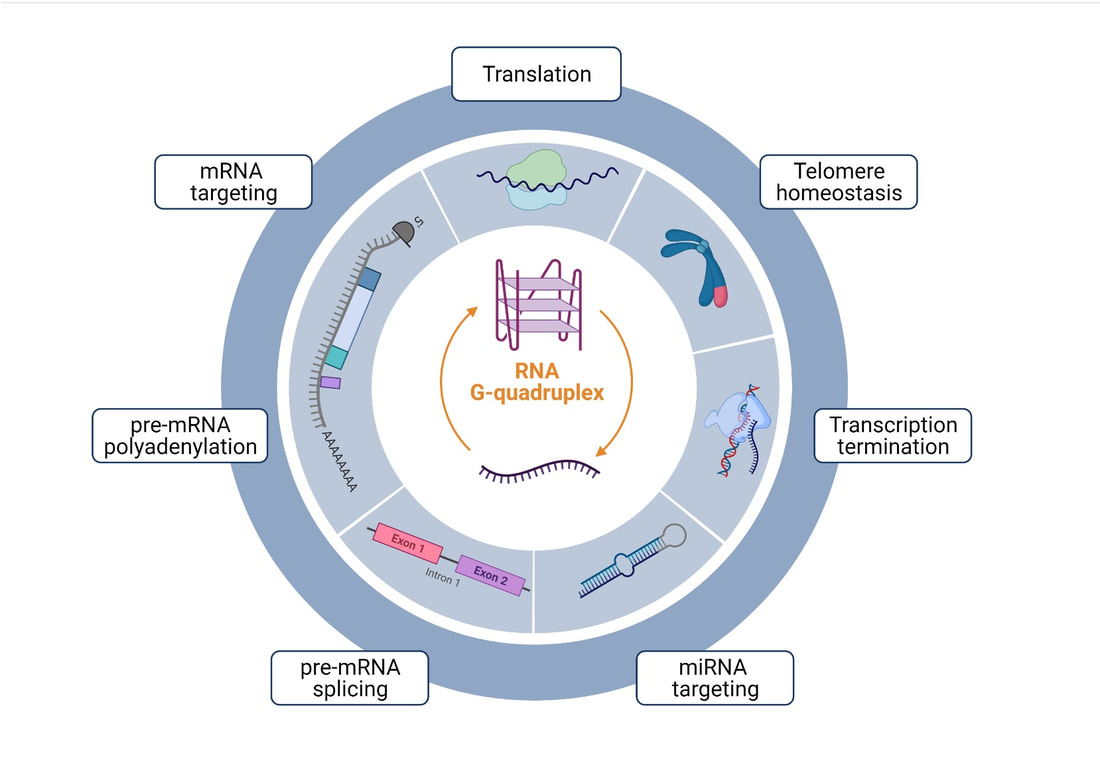
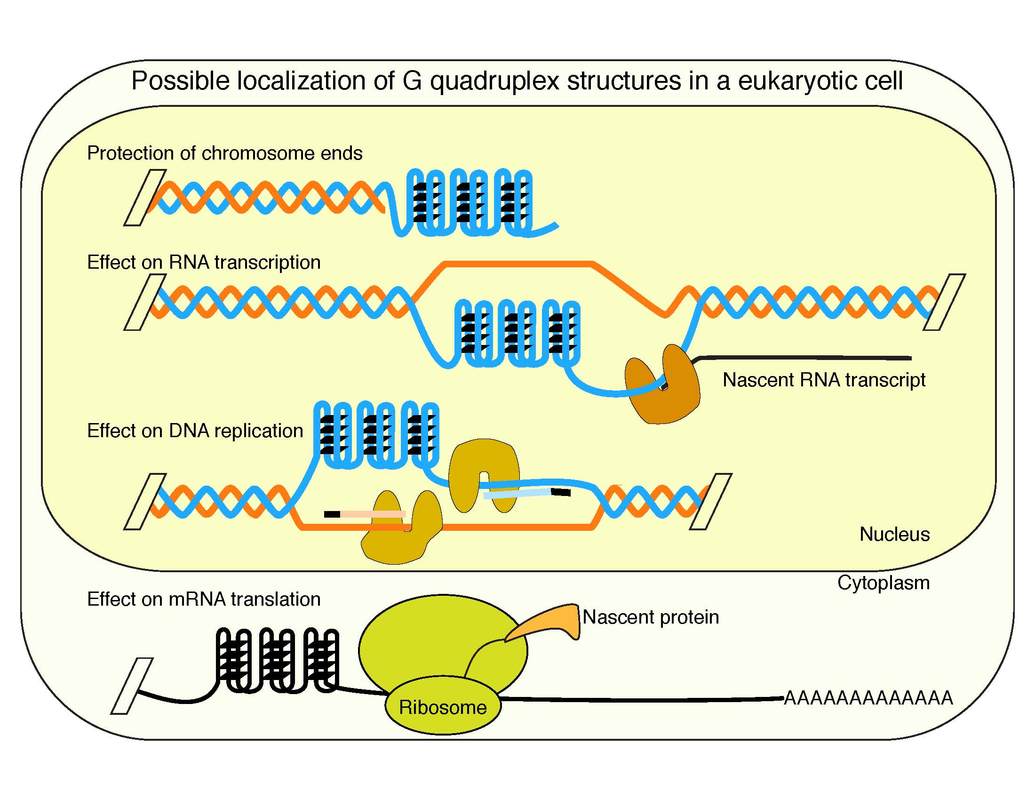
As G4 motif requirements for G4 structure formation are known, it was possible to identify the locations of G4 motifs in whole genomes using computational tools. They are enriched at specific regions such as telomeres, promoters, mitotic and meiotic double strand break (DSB) sites, and origins of replication (1-3). Accordingly, G4 structures could be involved in telomere maintenance, (post-)transcriptional regulation, or meiotic DSB formation (Figure 1a-e). To date, it is not known if G4 structures are relevant for one or several of these biological processes. Furthermore, the specific function of identified G4 structures in these processes is still not understood.
The current model is that from the 700.000 potential G4 structures, not all will form at once. Rather there will be a specific set of G4s that will form upon specific stimuli (e.g. infection, inflammation, stress, development etc) which support cellular processes, but can also contribute to genome instability. Therefore G4s are a double edge sword that needs a tight regulation by proteins. Research in the Paeschke lab has contributed significantly to the in vivo understanding of G4 structures and revealed how G4s affects positively telomere maintenance, DNA replication, and genome stability. We set the formation and unfolding of these structures into the context of cellular function during healthy and diseased stages. We focus on two major aspects that are impacted by changes in genome stability:
We addressed the question: How do G4 cause impact genome instability? which proteins are mutated or missing during these events? and which G-quadruplex DNA and RNA structures:genome stability - tumorigenesisgenome stability - immune response and changes within virus host interaction |
Research Projects: |
|
- Reveal the function and regulation of G4 structures during Nucleotide Excision Repair (NER)
- G4 - NER - potential connection to skin cancer development?
- Analyze the impact of DNA and RNA helicases and G4 formation during stress response
- Characterization of DHX36 function during viral stress
- Contribution of G4 structure for innate immune response
- How do G4 landscape changes alter during inflammation and how this triggers and immune response
- Determine telomerase function within the genome, contribution to genome stability and aging
- viral G4s how are they sensed by the host cell?